Details of the Drug
General Information of Drug (ID: DM0D9YJ)
| Drug Name |
Dirithromycin
|
|||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Dirithromycine; Dirithromycinum; Diritromicina; Divitross; Dynabac; Noriclan; Valodin; Dirithromycine [French]; Dirithromycinum [Latin]; Diritromicina [Spanish]; LY 237216; AS-E 136; ASE-136; Dynabac (TN); LY-237216; Dirithromycin (USP/INN); Dirithromycin [USAN:BAN:INN]; (9S)-9-deoxo-11-deoxy-9,11-[imino[2-(2-methoxyethoxy)ethylidene]erythromycin
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Indication |
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Therapeutic Class |
Antibiotics
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Affected Organisms |
Enteric bacteria and other eubacteria
|
|||||||||||||||||||||||||||||||||||||||||||||||
| ATC Code | ||||||||||||||||||||||||||||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Structure |
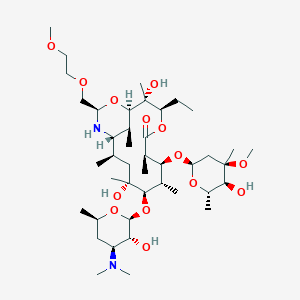 |
|||||||||||||||||||||||||||||||||||||||||||||||
| 3D MOL is unavailable | 2D MOL | |||||||||||||||||||||||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 3 | Molecular Weight (mw) | 835.1 | ||||||||||||||||||||||||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 4.2 | |||||||||||||||||||||||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 12 | |||||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 5 | |||||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 16 | |||||||||||||||||||||||||||||||||||||||||||||||
| ADMET Property |
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Identifiers |
|
|||||||||||||||||||||||||||||||||||||||||||||||
| Cross-matching ID | ||||||||||||||||||||||||||||||||||||||||||||||||
| Repurposed Drugs (RPD) | Click to Jump to the Detailed RPD Information of This Drug | |||||||||||||||||||||||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Transporter (DTP) |
|
||||||||||||||||||||||||||
 Drug-Metabolizing Enzyme (DME) |
|
||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | |||||||||||||||||||||||||||
References
